Hi everyone! Erin, Embark’s Senior Veterinary Geneticist, here.
As more people have been choosing Embark for their genetic testing needs, I’ve been receiving quite a few questions about our technology — what is “DNA microarray technology?” “Is it proprietary?” “How do you know it’s accurate?” “How is it different from the single mutation tests that I can purchase from other services, and why don’t you offer single mutation testing?” I thought I might spend a bit of time explaining the differences between the two.
Here’s a comparison:
What is the molecular method?
Single Mutation Test: Amplifies and evaluates DNA sequence at a single genomic region
Microarray Genotyping: Uses probes to sample regions across the entire genome (density, aka, probes per stretch of sequence varies by service)
What kind of results do you expect?
Single Mutation Test: Your dog’s genotype at a single location
Microarray Genotyping: Your dog’s genotypes at thousands of locations in the genome, providing breed ancestry, sex, relatedness, as well as health and traits
Can you purchase tests à la carte?
Single Mutation Test: Yes
Microarray Genotyping: No
What is the expected turnaround time?
Single Mutation Test: Several days to several weeks
Microarray Genotyping: 2 to 4 weeks
Are the methods proprietary?
Single Mutation Test: Sometimes, but most scientifically validated assays are published after peer-review
Microarray Genotyping: Most of Embark’s content is from the well-established Illumina CanineHD array, developed by the Broad Institute and others. Embark has also added custom probes for health and traits that you can’t buy online. Other services’ microarray technologies are completely custom and unavailable online.
What kind of quality control is in place?
Single Mutation Test: Most services train their assays on known positive controls. Ideally labs use two independent assays to ensure that the results agree. However, they have generally no way to detect sample mix-ups or confirm the appropriate sample was sent by genetic means. In addition, the exactly quality control protocols will depend on the service.
Microarray Genotyping: As above, we train our probes on known positive controls. Embark uses multiple probes per mutation to ensure inter-probe agreement, and our genotyping is performed at a very large, CLIA-certified lab with effective and standardized procedure. Our geneticists oversee results and can run single mutation tests to confirm unexpected results. Finally, Embark’s high-density genotyping and policies can confirm sample identity through genetic breed, sex, and relatedness/parentage of any sample.
What services use which method?
Single Mutation Test: Paw Print Genetics, Gensol Diagnostics, Laboklin
Microarray Genotyping:Embark Veterinary, Genoscoper Labs
SINGLE MUTATION TESTING
Most services that offer single mutation testing use classic molecular biology techniques such as Polymerase Chain Reaction (PCR). PCR revolutionized the way scientists evaluated DNA. Instead of needing massive amounts of DNA to identify a sequence (literally, we are talking gallons of saliva instead of just a cheek swab), PCR allows us to amplify a small amount of DNA exponentially. The developer of this assay, Kary Mullis, actually won a Nobel Prize for this contribution to science!
Here’s a very basic diagram that describes what exactly we’re doing with PCR. In brief, PCR uses heat to “unwind” the DNA double helix (word for the wise, don’t boil yourself! Your DNA will unwind, among other things), then rapidly cooling it to allow short DNA fragments called “primers” to bind the DNA. These primers are designed to specifically bind a region of the genome, usually flanking a mutation of interest (which I’ve delineated with a yellow dot). The temperature is then adjusted to allow an enzyme called Taq polymerase to start making new DNA complementary to the unwound DNA template, using our primers as a starting point. Just with one cycle, then, you’ve doubled the number of DNA copies. If you do it another 30-40 times, you’re increasing the number of DNA molecules in your reaction 2^30 or 2^40 times.

Or if you prefer a video, click here.
Downstream analyses of PCR products can include Sanger sequencing or fragment analysis. Both of these methods assay the base composition or the size of the PCR product. Depending on the nature of the mutation you’re looking at, you may choose one over the other. Click here for a nice video.
Another iteration of a single mutation assay uses fluorescent probes for allele-specific PCR, like the TaqMan assay.
The TaqMan assay also uses primers, as in normal PCR, but uses fluorescent dyes that can indicate which version of a SNP (allele) is being amplified by the reaction, and can give insight into how many copies of the allele a dog has (quantitative PCR).
Those of you with especially quick minds may already see one huge drawback to using PCR. Contamination is an ISSUE. If you’re taking a single copy of DNA and amplifying it quite literally 2^40 times (so that one copy goes to about 109,951,160,000 copies in about two hours), the possibility of another dog’s DNA sample sneaking in and also getting amplified is entirely possible. As such, stringent methods to control for PCR contamination are compulsory. Click here for an article that gives a sneak peek into what lengths molecular biologists go to minimize contamination.
In addition, we all know that random genomic variation occurs across dogs. Primers are designed to bind a specific region of the genome based on 100 percent unique sequence that maps to that region. We usually design our primers off the reference genome, which was constructed from a Boxer named Tasha. However, if a dog happens to have a mutation within the sequence that the primer is supposed to bind, it’s possible that the primer won’t bind at all, leading to a false negative result. For example, a mutation in the SOD1 gene, associated with degenerative myelopathy (DM), was interfering with primer binding and leading to false negatives in routine PCR-based DM testing–potentially a devastating result for breeders of high-risk breeds. So, along with the standard measures that are taken to control for contamination and ensure test accuracy, most single-mutation testing services address the possibility of a single test failure by applying at least two different assays (let’s say, two different primers pairs, or fragment analysis AND Sanger sequencing, or TaqMan AND Sanger sequencing) to increase confidence in the genetic result.
The kicker, though, is that even with measures like the above, the data you glean from a well-done PCR-based assay is that you’re 100 percent sure of the dog’s result at the genomic region of interest, and only that region. You glean no further data from this sample, meaning that if a different test needs to be done, you have to run another assay. And you cannot identify the sample by genetic means. In other words, to pair the genetic result to the actual dog relies on accurate documentation by the client–reporting sex, breed, parentage, etc. is not linked to the dog’s genetic result, making the results subject to intentional or accidental manipulation or sample mix-up.
In short, all of these assays in general will give you high quality information on a single mutation. If you’re buying a battery of tests, you might get information on the eight to 20 that you select to test your dog on. But you do not in general get additional information that can raise your confidence that you’ve tested the right dog.
MICROARRAY BASED GENOTYPING
Microarray technology are a well-established technology used to assay hundreds of thousands of mutations at once. Often referred to as “SNP chips,” “SNP” stands for single nucleotide polymorphism, a particularly informative type of genetic mutation. SNP chips have been used for over 15 years and are the standard for high-quality direct-to-consumer human DNA tests (the FDA approves of their use for testing human genetic conditions, and this is the technology that 23andMe, Ancestry, and every other human direct-to-consumer DNA test uses). They are also the standard for scientists finding new health conditions and are widely used for agricultural purposes.
Embark’s SNP chip is based on Illumina’s CanineHD SNP array, assays over 200,000 genetic variants that are known to occur in different dog breeds. The base technology is not proprietary — you can actually buy CanineHD chips right off the shelf, if you like, as dozens of scientists do every year. We’ve just added tens of thousands of custom probes that inform Embark-specific features like coat traits, health risk, and wolfiness.
Take a spin through the technology here.
And for those who want to get into the nitty gritty of the actual protocol, here’s a great resource.
As you can see in the videos, microarray-based genotyping uses synthetic DNA probes labelled with fluorescent chemicals. If a dog’s DNA binds a specific probe, the chemical fluoresces at a specific wavelength, or color. As such, microarrays define genotype based on the intensity and wavelength of the probes that bind to a dog’s DNA by the nature of DNA to seek complementary sequence. So quite literally we are evaluating what color your dog’s DNA is fluorescing, and since we know what color we’ve assigned to which genetic variant, we infer what your dog’s genotype is based on the color.
A lot of the questions I get center around accuracy. For many people, it’s easier to understand how a PCR-based result can be accurate compared to how a microarray-based result can be accurate, possibly because people are much more familiar with PCR. Hopefully I’ve already upped your familiarity with microarrays, but here are the specific measures we use to make sure our results are as accurate as can be (well over 99.9 percent accurate).
- For rare mutations, we train our probes on known heterozygotes and homozygotes. We very commonly offer complimentary tests for dogs who have known genotypes for rare disease risk alleles. This is exactly what other services do when they ask for known carriers or homozygotes–they want to train their assays on positive controls, too. It’s just that their assays are different, as I described above.
- We design several probes that query the same mutation. If two, three, even eight probes are all in agreement for a genotype, our confidence is doubled, tripled, octupled that the result is right! And, if probes do not agree, we manually review and see what’s going on. Usually, this means one probe is misbehaving–we then exclude that probe from analysis.
- Similarly, we test a number of dogs several times on different chips. When we upgrade to a new chip, we make sure our probes call the same dog the same way every time.
- We regularly subject our results to manual review and validation. Whether this is by manual review of probe data OR by testing a dog in-house via those classic molecular techniques, PCR, sequencing, or fragment analysis as I discuss above, should their genetic result be inconsistent with what you know about the dog. Please email us at [email protected] if you have concerns! Recently, we caught a misbehaving probe for the dilute trait, D Locus, that incorrectly called some “DD” dogs as “Dd.” With the help of some of our breeders, we quickly identified this issue, and corrected the very small number of dogs affected.
- We confirm that this sample belongs to the dog you’re testing. Because we have 200K probes that inform ancestry, sex, and appearance, we can confirm that this is not an issue of sample misidentification (and sometimes breeders do accidentally assign the wrong swab to the wrong profile!). Further, if a dog is cleared by parentage, we are happy to test the parents to confirm their genotypes AND the parent-offspring relationship. We have caught a few unexpected sires this way!
- We can detect contamination at the start of our analysis. We can use microarray data to identify sample contamination simply because, unless a dog is a chimera or the cells samples are polypoid (normal in some fish, amphibians, and plants, abnormal in dogs), a dog really should only have at most two different alleles at a given SNP (because they should only have two copies of their genome, one from mom, and one from dad). Two dogs will inevitably differ in sequences at least a few probes, meaning that if a sample is contaminated, some probes are going to report three, maybe four alleles. Our pipeline catches these reports, fails the sample immediately on the basis that there is probably contamination, and we issue you a new swab for the dog.
Now that we’ve laid those out, I’m going to go into some nitty gritty details of actually using probe binding intensity to infer genotype.
A LITTLE MORE ABOUT INTENSITY-BASED GENOTYPING
At the nano-scale level, if we consider a single probe, stuck to just one bead, embedded into our silicon wafer, this is essentially what we’re looking at:

For a simple recessive mutation like the mutation that is associated with PRA-prcd, I would then label the “Homozygous mutant,” “Heterozygous,” and “Homozygous wildtype” genotypes as “At Risk,” “Carrier,” and “Clear.”
But remember, our SNP chips have tons of probes, which will make tons of fluorescent dots. So really what your dog’s run looks like is more something like this:
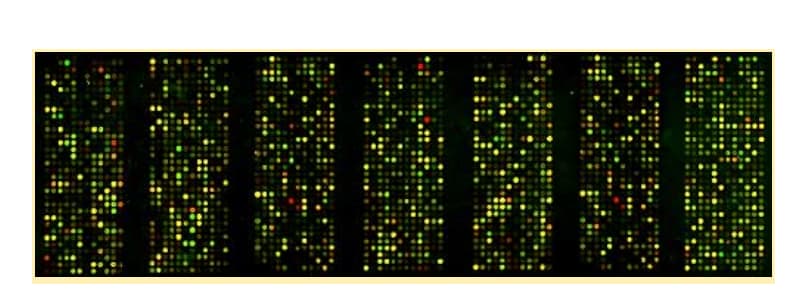
Courtesy of the University of Washington School of Medicine.
You can appreciate even in this image that some of these probes don’t look exactly the same. Some are “brighter” than others (variations in intensity). Some of the yellow dots skew more towards orange or lime green than others (variations in wavelength). This is simply due to the fact that these probes vary in sequence. Let’s take a quick dive into how exactly, then, we evaluate this variation and ensure accurate, replicable results.
When we’re talking about evaluation, we are generally evaluating patterns in probe binding variation across LOTS of dogs. Remember–we’re rarely running just one dog at a time. And one pooch does not a pattern make. Instead, we test thousands of dogs at the same time, and we’re evaluating their probe results all at once. Below are some simplified examples of plots that reflect actual probes on our chip. Each plot reflects probe patterns for somewhere between 1,000 and 12,000 dogs (just depending on what window I had open). On these plots, the Y-axis always measures intensity or brightness of fluorescence; the X-axis measures wavelength (color).
In a perfect world, our probes are going to look like this, even if we’re evaluating tens of thousands of dogs at once. All of the dogs’ intensities are sitting right on top of each other–even though the rightmost cluster has over 11,000 dogs, we can clearly see that they all have the major allele, whereas the three all the way to the left obviously have the minor allele. This probe also performs admirably as far as wavelength: You can see that the middle cluster of dogs sits right in the middle of the left and right cluster–that is, heterozygotes are fluorescing at an intermediate wavelength relative to the homozygous clusters.

Of course, not all probes perform the same. Some of this is sequence dependent; some is batch-dependent. This probe is a little less pretty, you can see that these dogs are spreading out a little more wavelength-wise, making visibly “larger” clusters. But we can still clearly evaluate which dogs have what genotype.

In addition, some probes don’t have the exact wavelength pattern where the heterozygous cluster is exactly halfway between the two homozygous clusters, which you can appreciate above. Some have wavelength patterns that are slightly shifted, for example:

As you can see, the heterozygous cluster in this image is actually shifted quite a ways to the left. But it’s still very clearly a cluster. So as long as we know that this is the way this probe acts due to manual review of our probe repertoire, we can adjust our cluster positions for this probe, thereby controlling for its idiosyncrasies. Cluster updates happen on a regular basis here at Embark as we monitor the data coming in.
In addition, some probes are also going to actually have a bit of a “tilt” to their cluster.

Above, you can see that the heterozygous and homozygous “wild type” clusters both have a bit of a tilt as well as a tail. Depending on where we’ve delineated our confidence intervals, some of these dogs might receive a “No Call” at this probe. Again, this is totally okay for two reasons. First, because we design multiple probes for each mutation of interest, even if one probe is a “No Call,” the other probes should allow us to provide a result. And if for some reason a few probes are throwing “No Calls” due to a “tilt” happening with several probes, leading to an overall “No Call” result, it will be flagged for manual review and corrected.
Note that, while a lot of this curation is done before you even see the results, we do always want you to point out any concerns. If a result ever comes into question, please reach out to us at [email protected] and we will do our best to address it.
Hopefully, this has been a nice walk through some of the different methodologies employed by different DNA testing services, as well as the steps that we take at Embark to ensure that your results are as accurate as they can be. We take a lot of pride in our testing. We, like you, value affordable and accurate genetic testing. Additionally, our platform provides unprecedented potential to learn more about your dogs: those 200,000 markers are not just informative for ancestry and relatedness, they are the fuel for future discovery! We are never far away from the individual dog: our scientists spend time every day looking at individual samples, and in communication with our users. And if you haven’t noticed, we love talking about science–please do not hesitate to reach out if you want to learn more about what we do.













