Many dog breeders have reached out to us to ask “What makes Embark’s test better for maintaining diversity and lowering disease risk in my breed?” Specifically, they are often interested in how Embark compares to other diversity management products that use a very small number of genetic markers to make breeding recommendations.
In short, you cannot assess and preserve diversity in regions of the genome that you do not analyze. In fact, using a very small number of genetic markers to inform breeding decisions may be harmful to genetic diversity. I know this may seem counterintuitive, but as the old saying goes, the proof of the pudding is in the eating… So to demonstrate the risks of using low-density marker panels for preserving diversity we set up virtual breeding simulations to compare several alternative approaches to diversity preservation.
For this experiment, we used genetic simulation software (SLiM) to simulate a simple model of the dog genome and 20 generations of breeding management in a single dog population/breed. We simulated haplotype diversity, as we would measure with Embark’s genotyping array, AND diversity at sparsely positioned genetic loci, specifically 33 loci positioned across the genome in a manner similar to UC Davis’ STR panel. We replicated each simulation 100 times to explore random variation in each model. In addition to a totally random mating model, we examined a few different methods for choosing optimal mates to preserve genetic diversity, defined here as average allelic richness, or the average number of unique stretches of DNA in each region of the genome in a population:
- Pedigree: This model avoids inbreeding between very close relatives based on a two generation pedigree (back to grandparents).
- Genome-Wide Heterozygosity: This model chooses mates that maximize average haplotype heterozygosity across 50 kilobase haplotypes as we might measure using Embark’s genotyping array.
- Genome-Wide Relatedness: This model chooses as mates the most distantly related dogs in a population as inferred from shared 50 kilobase haplotypes, like those calculated by Embark’s COI and expected COI scores.
- STR Heterozygosity: This model chooses mates that maximize average per-site heterozygosity across the 33 STR markers in offspring.
- IR: This model chooses mates that minimize Internal Relatedness (Amos et al 2001) across the 33 STR markers in offspring.
Figure 1: STR-based mate choice leads to faster loss of genetic diversity than random mating

As you can see in Figure 1, significantly more genetic variation is lost when we only use the 33 STR positions to guide breed management (in all comparisons of an STR model to any other model P < 10-48 with a 2-sided Mann-Whitney U test). This is because dogs with rare alleles at one or a few markers may be strongly favored as parents for the next generation regardless of the genetic variation throughout the rest of their genome. This “popular parent” effect causes diversity loss to accelerate because it misses the > 98% of the genome not tagged by STR markers.
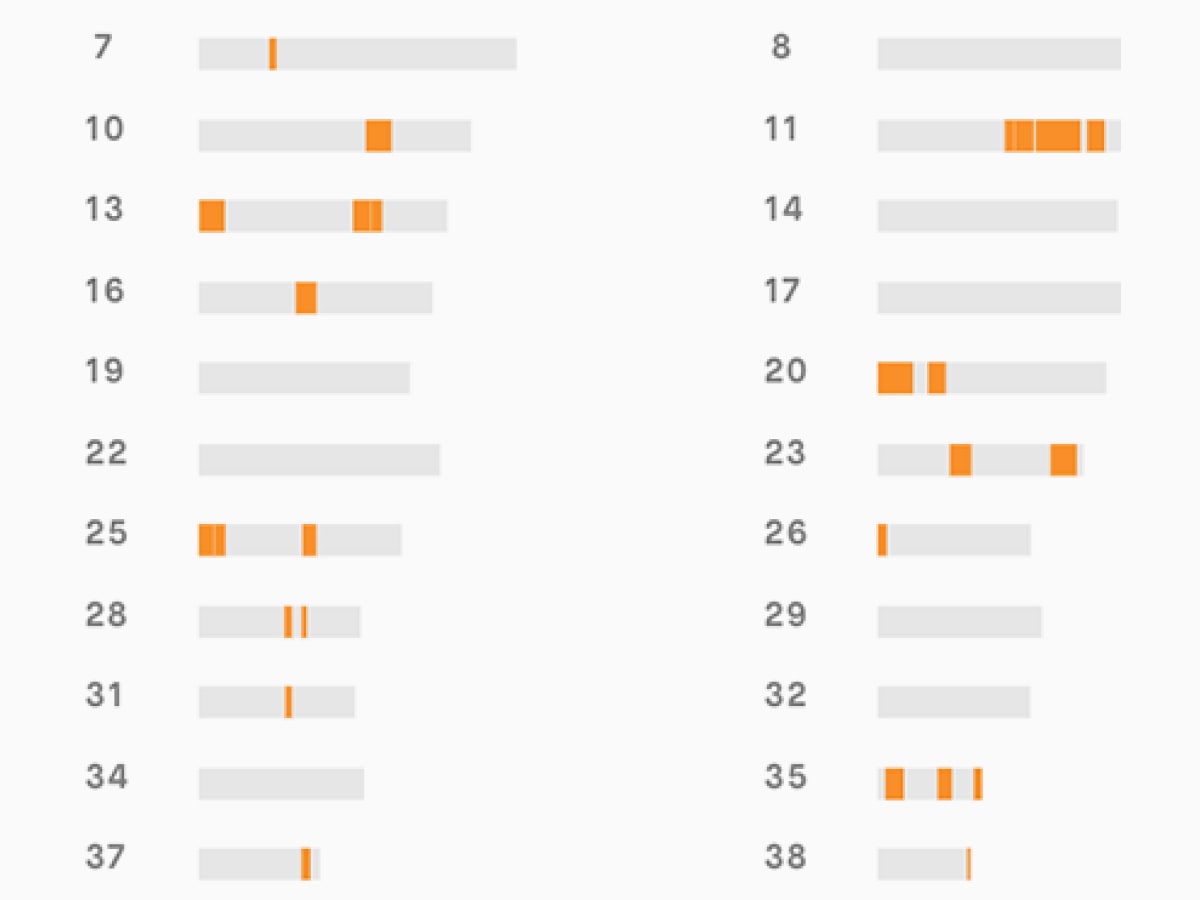
Figure 2: STR-based diversity management works well near STRs but very poorly away from STRs

This effect is very apparent if we look at how diversity is lost in relation to proximity to STR markers. You can see in Figure 2 that with STR-based mating schemes diversity is preserved very well at, and very near to, STRs but is quickly lost as you move away from those markers. 98.5% of the genome is more than 500kb from one of the tested STRs. If the goal is to preserve diversity, it is actually worse to use these STRs than to select mating pairs at random.
If you don’t have a pedigree, as is usually the case in conservation genetics of wild populations or wild-caught individuals, then the use of a scattering of SNPs or microsatellites to determine who is closely related can be useful to manage inbreeding. Additionally, low-density marker panels can be used to determine if there are genetic differences between populations that need to be taken into account during the breeding program, but their utility is limited.
You may hear that statistics like Internal Relatedness (IR) calculated on low-density marker panels are used all of the time in modern scientific studies. This is true. However, it is very important to keep in mind that even though some statistics like IR and others were designed by scientists to take a snapshot of genetic diversity in a population within a single generation using a limited amount of genetic data, this methodology was not designed to track and maintain diversity in a population over time. In fact, many previous scientific studies have shown that estimates of heterozygosity from low-density marker panels are very ineffective at capturing signals of inbreeding– see this early study, and this much more recent study for just two such examples. Low-density marker panels are ineffective as estimators of variation for diversity maintenance, which requires accurate estimation of inbreeding. In the absence of high-density genome-wide analytic tools, you are better off using pedigrees. Estimate relatedness in the founder generation, then maintain your pedigree information (in combination with paternity testing) for long-term breed management. It will be more accurate than a low-density marker panel.
We know that it may come as a surprise that using a limited amount of genetic data for diversity management could be worse than using none at all, but that is what science supports. At Embark, we are working to develop methods to take genome-wide genetic information and apply these data in a way that specifically preserves diversity. Our results above are promising, but to properly manage diversity in these populations requires more research, including simulating the effects of various test statistics and breeding scenarios on the population, before releasing a test.
So what can you do now to maintain health and preserve your breed’s genetic diversity? Use the best tools available to avoid unnecessary inbreeding, test for mutations that are known to associate with health risk, use results as well as your knowledge of your lines to avoid perpetuating a disease in your breed, and keep as many breeding animals of both sexes in the gene pool as possible.
How is Embark supporting this effort? Embark’s current test offerings focus on genome-wide panel testing of known trait and disease variants and accurate inbreeding and relatedness coefficients. Furthermore, generating a digital DNA database for a breed over 200,000+ markers helps research efforts for both diversity analysis and for trait and disease mapping– a platform that accelerates new discoveries to provide breeders with even more informative genetic tests and actionable breeding tools in the future.
We know that this is a lot to consider at once, and we certainly wish that there was a genetic test immediately available that could work more directly to improve genetic diversity, especially for so many breeds that are currently at great genetic peril. Embark and other researchers are doing our best to develop more efficient methods to preserve breed-wide diversity. However, while we are all working together to develop tools to help improve the genetic health of your breed, there is a lot that you can do now with scientifically validated tests, your own knowledge of the breed, and expert assistance from population geneticists and veterinarians.
Embark is on a journey to improve canine health. We, just like you, want what is best for your dogs. We hope you will join us.
*Many thanks to Brett Ford, Bioinformatics Research Associate at Embark, for designing these simulations and Adam Boyko, Chief Science Officer at Embark, and many others for their contributions to this article.